Select analysis version to view the applicable content:
Last time we discussed what an artifact is, why it is important to correct artifact, and how artifact is identified in the MindWare analysis applications. The next step is actually dealing with that artifact. We are going to start with some examples which, while they may seem obvious, are important to understand before proceeding to the more complex scenarios. Beyond highlighting some common artifact seen in ECG data, these examples will also speak to the features available in the ECG Editor.
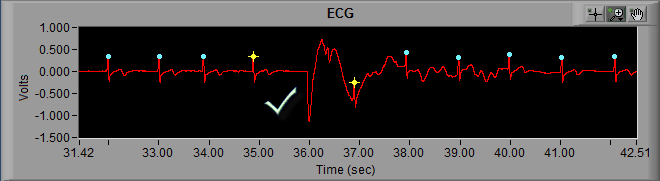
Adding a Missing R Peak (Visible in Data)
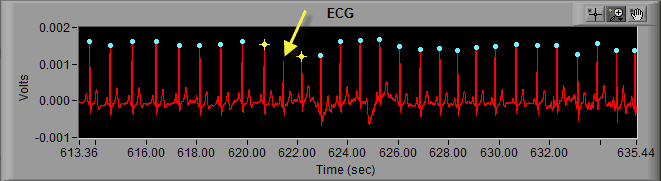
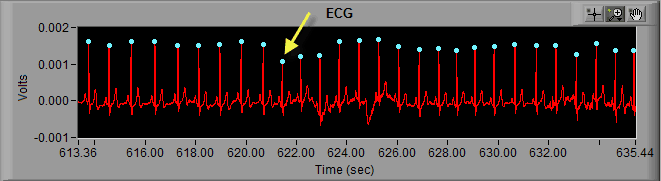
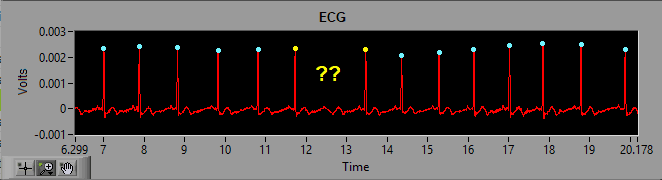
As we discussed previously, the dynamic R peak detection algorithm attempts to identify R peaks based on known characteristics and morphology of ECG. Occasionally, if a peak does not meet this criteria, it will not be marked as an R peak.
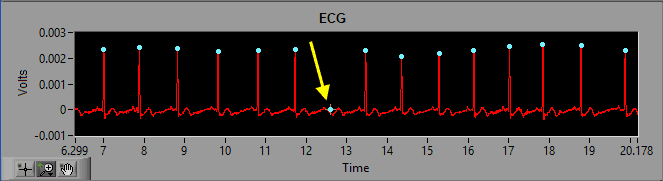
As we can see in the above screenshot the highlighted R peak has not been marked and, due to the resulting elongated IBI, the beats around the missed R peak have been flagged as artifact. But by looking at it we can clearly see a normal cycle of ECG which has simply been missed by the peak detector. The solution here is to simply insert that peak on the ECG waveform.
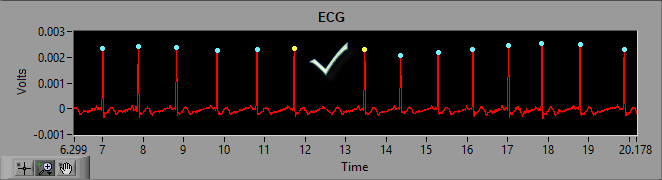
Deleting an Erroneous R Peak
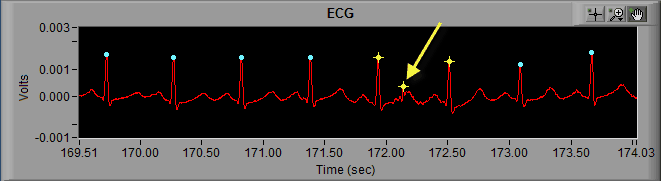
Much in the same way that the peak detection algorithm can miss an R peak, it can also falsely identify noise in a signal as an R peak.
Here we can see noise in the ECG signal which meets enough of the characteristics of an R peak that it is being marked as such. Visually, we can clearly see that it isn’t an R peak. Not only is the amplitude far too small, but it also represents a physiologically impossible change in IBI. Hence the artifact detection algorithm marking it and the peaks around it with yellow stars. Once the beat has been determined to be improperly marked, the next action is to delete it.
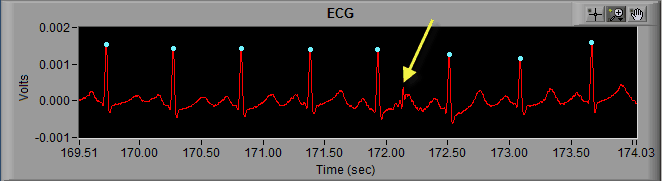
Once the peak has been removed, proper timing has been restored and the segment is clean.
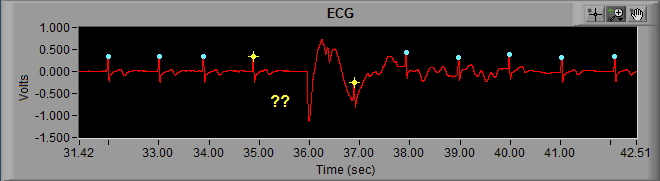
Adding a Missing R Peak (Not Visible in Data)
This all seems pretty easy so far, right? In each of the last 2 examples, it has been obvious where the R peaks exist, so it is pretty straight forward to correct any mistakes the peak detection algorithm has made. The common theme is that we have been working with a good quality ECG signal. The reality is the ECG signal will not always look this nice, and it may be difficult (or impossible) to determine the location of the R peak.
To deal with this situation, we first need to consider what type of analysis we are performing:
Heart Rate Variability
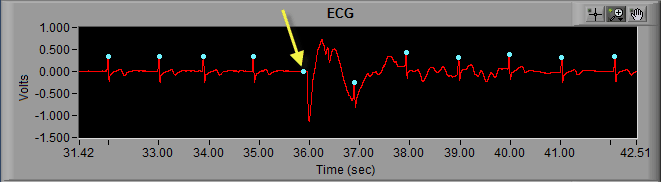
With HRV, we need to preserve a contiguous R peak series (of at least 30 seconds) to calculate RSA. When we can’t visually identify an R peak in the signal, the best we can do is estimate its location. This is done by splitting the inter-beat interval down the middle, thus creating consecutive IBI’s of the same value (and introducing minimal skew in variability). In the MindWare ECG Editor, this is known as a Midbeat.
Impedance Cardiography
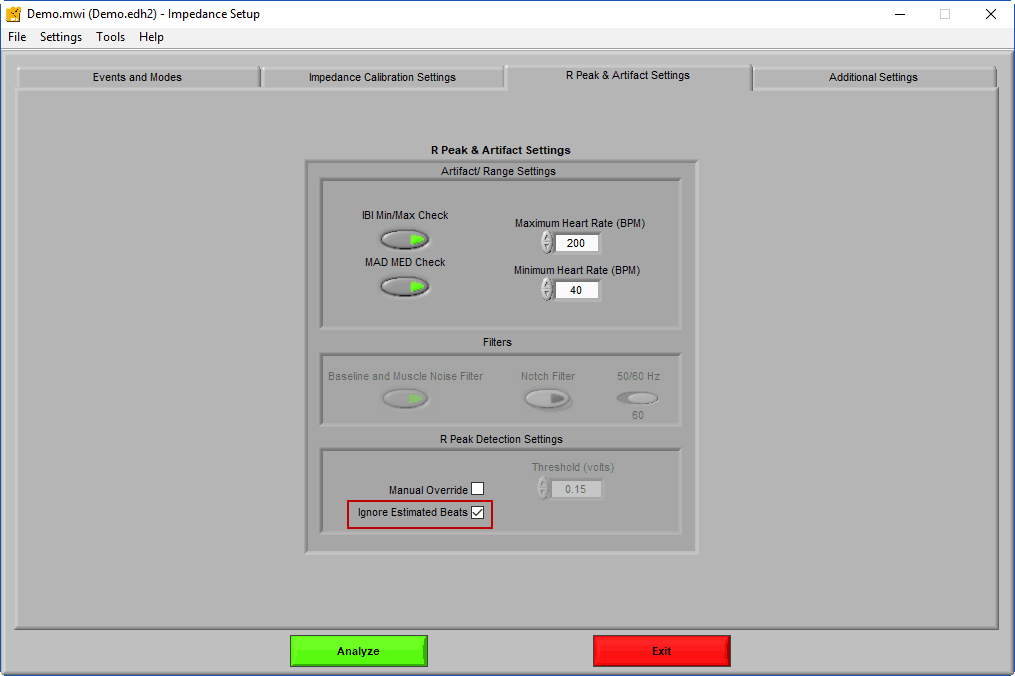
For impedance cardiography, since we mainly care about good quality cycles of ECG with which to build our ensemble waveform, we actually would not want to do anything to the data. The MindWare IMP application automatically ignores artifact-based IBI’s when calculating heart rate to allow for discontinuities in the series of R peaks.
Note: By enabling Ignore Estimated Beats on the IMP Setup screen, midbeats will be excluded from the Ensemble Average, thus improving re-usability between HRV and IMP edits
10% Rule
If the R peak is visible in the ECG waveform, you can insert/delete as many as you need! So long as you can visually identify the R peak then no information is lost. However, when performing the Midbeat, information is lost in the process of IBI estimation. Too much of this can skew the final results, which we don’t want to happen. As a rule of thumb:
It is best to keep the number of estimated beats under 5% of total R peaks in a given time segment, but is probably OK up to 10% of total R peaks.
Any more than this and it will become difficult to justify results in a publication. Reminder – this concept of estimating R peaks primarily pertains to HRV analysis.
If you find yourself needing to estimate more than 10% of the R peaks:
In a given segment, you should drop the segment from analysis. If you at least have a 30-second section of ECG in which R peaks are clearly identifiable, you can possibly delete problematic sections and salvage the segment.
In a given epoch, you should drop the epoch. Design your study with replicate epochs to allow for some to be discarded without affecting the end results.
In a given subject, you should drop the subject. It is unfortunate when this happens, but it is important to recognize when this needs to be done. Report the reasoning for dropping the subject, and move on.
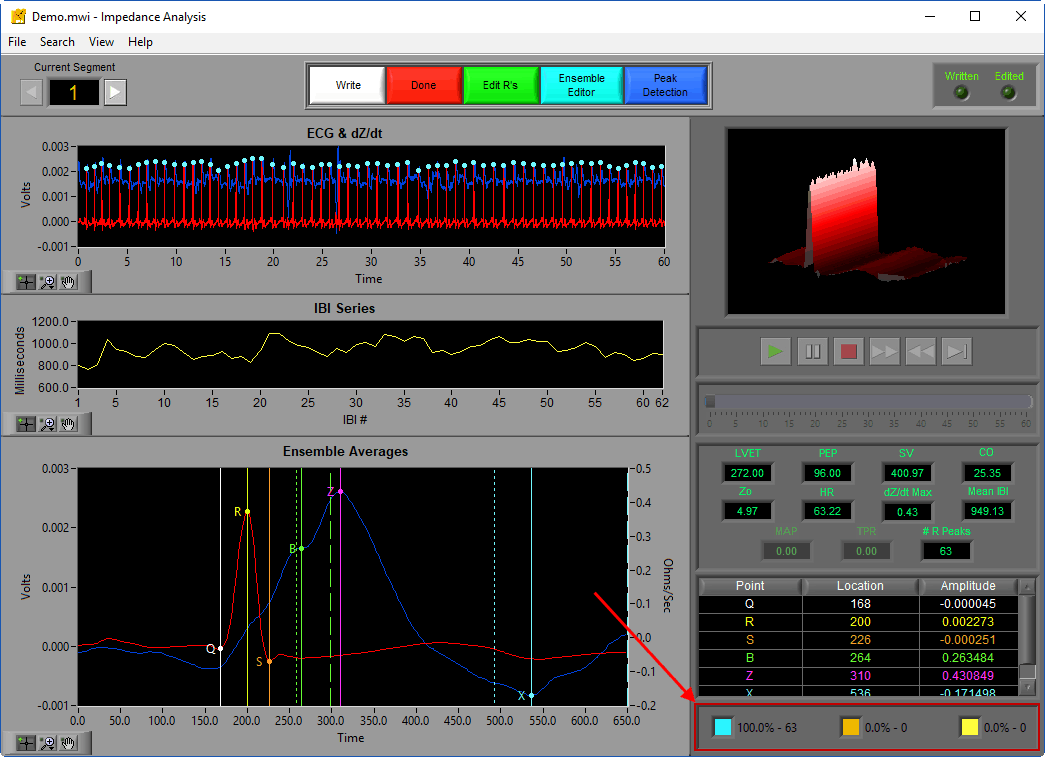
Take advantage of the Point Distribution editing statistics to ensure that you are staying within these limitations, paying particular attention to Estimate peaks marked orange.
Onward and Upward
Hopefully at this point you have a basic understanding of how to clean an ECG series in some common scenarios. Next time we will start looking at more complicated instances of heart conditions and arrhythmia.