Select analysis version to modify content below:
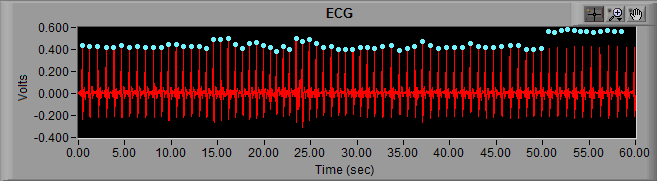
Symptom: Blue peaks roughly align with R peaks on ECG signal, but appear to hover above or below actual peak.
Note: In version 3.2 and greater, this cannot occur because channel mapping and filtering settings are automatically applied by the edit file
This is most likely due to a change in filter setting since the data was originally edited. The edit files do not contain filtering information, so it is imperative that the current filter settings match the settings at the time of editing
Solution:
- Check the Baseline & Muscle Noise Filter and Notch Filter settings on the R Peak & Artifact Settings tab
- Check the BioLab Filter setting by going to File>>Scaling & Filtering Settings
- As a rule, if the peaks float above the signal then there are filters applied currently that weren’t applied when the data was edited (and vice versa)
Symptom: Peaks appear erratic, and unrelated to the ECG signal
This is typically seen when the incorrect edit file is loaded for a given data file or subject.
Solution:
- If you have multiple subjects in your data file, ensure that the correct edit file is being used for the subject being analyzed. Each edit file can only contain a single subject’s data
- Check to make sure the edit file that has been mapped matches the data file being analyzed
Symptom: Peaks appear erratic, and unrelated to the ECG signal
This is typically seen when the incorrect edit file is loaded for a given data file or subject.
Solution: Make sure the edit file selected matches the currently opened data file