HRV vs. IMP : Why R We Editing Twice?
Collecting and editing the ECG signal is crucial to performing heart rate variability (HRV) and impedance cardiography (IMP) analysis. In both of these cases it is necessary to identify a landmark on the ECG waveform to identify a beat. Using a lead-2 electrode configuration, the most easily-identifiable landmark is the R peak. If both types of analysis require identifying the same landmark on the same signal, why can’t we always use the same edited series for both?
HRV – Measuring Variability
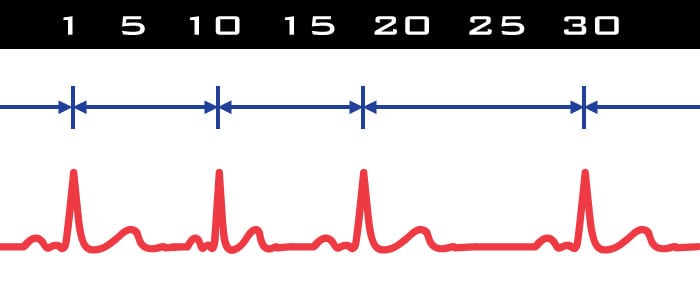
In HRV we are quantifying the variability of the inter-beat interval (IBI) over a specified time segment. Timing between beats is everything, hence it is crucial that R peaks are properly identified so the IBI is correctly determined. Take, for example, a section of data in which an R peak is unidentified or not present. The effect on the IBI series is a physiologically impossible change in IBI which will have an enormous effect on RSA. It is for this reason (and a few other more complicated reasons) that there must be a contiguous R peak series in HRV to calculate the beat-to-beat variability. Using the spectral method to obtain RSA imposes even more restriction on editing: at least 30 seconds of contiguous data is required to provide the Fast Fourier Transform (FFT) with sufficient information.
IMP – Ensemble Averaging
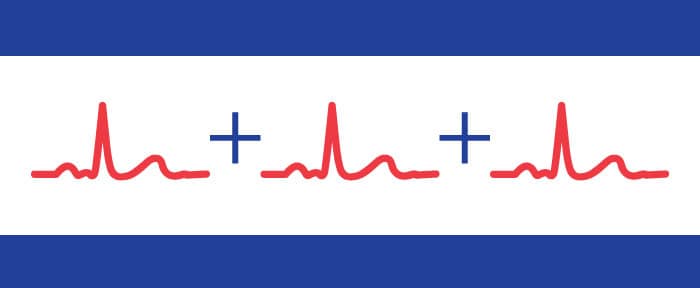
When we perform impedance cardiography analysis, we are not examining the timing between R peaks, but rather the timing between the electrical and mechanical activation of the heart found by comparing the ECG and dZ/dt waveforms. To do this, we create an ensemble average of a portion of each of these signals surrounding all identified R peaks. The key points on these aggregate waveforms (Q,R,S,T for ECG, and B,Z,X for dZ/dt) are then compared for timing statistics such as LVET and PEP. The ensemble will only contain identified R peaks – any which that are not identified or are missing will have minimal effect on the resulting ensemble waveform.
Can we ever reuse our edited ECG signal?
In an ideal world, the ECG signal is clean enough to allow for all R peaks to be identified, in which case it is perfectly acceptable to use the same edited ECG series in both heart rate variability and impedance cardiography analysis. Unfortunately we do not live in an ideal world and data is rarely perfect. There are cases where an edit needs to be made that renders the ECG unusable in HRV due to the non-contiguous R peak series, but it is completely valid in IMP. On the other hand there are times when the latter half of the ECG waveform needs to be discarded to make it valid in HRV, but reusing that same edit in IMP would be throwing away information which could have influenced the ensemble waveform. There needs to be careful consideration in determining whether using an edited segment in both HRV and IMP is advantageous, detrimental, or even acceptable before reusing the same edits for both types of analysis.
Back to the ideal world…
We may not live in an ideal world, but we can strive to get closer to one by considering a few key concepts:
- Proper training and a firm grasp on site preparation, electrode placement, and proper signal morphology can go a long way toward achieving the elusive “clean” ECG signal
- Collect your data with equipment that is best suited for your line of research
- When editing is unavoidable, understand how to edit for the type of analysis you plan to do and use tools to make the task of editing easier